Multivariate Normals
Published:
- Multivariate Copula Modeling
This R markdown is for explaining simple copula modeling in stan.
Table of content
Packages and setup
packages = c("brms","tidyverse","bayesplot",
"pracma","here", "patchwork",
"posterior","HDInterval",
"loo", "furrr","cmdstanr","mnormt")
do.call(pacman::p_load, as.list(packages))
knitr::opts_chunk$set(echo = TRUE)
register_knitr_engine(override = FALSE)
set.seed(123)
Overview.
In this markdown the goals is to build up an understanding of multivariate distributions. I will introduce this with Gaussian random variables. In this markdown I will show that there are 3 different ways of simulating such that two of which are practically equvient whereas the last is just a special case of the two. The three types of models are:
Two marginal Gaussian (no off diagonal in the variance covariance matrix)
A multivariate Gaussian with a mean vector and a variance covariance matrix
Two marginal Gaussian with a gaussian copula linking the two marginals
Forward simulations
Two Marginal Gaussian Distributions without a variance covariance matrix
Two independent univariate Gaussian random variables can be written as:
That is,
\[\mathbf{Y} = \begin{bmatrix} Y_1 \\ Y_2 \end{bmatrix} \sim \mathcal{N} \left( \begin{bmatrix} \mu_1 \\ \mu_2 \end{bmatrix}, \begin{pmatrix} \sigma_1^2 & 0 \\ 0 & \sigma_2^2 \end{pmatrix} \right)\]- A multivariate Gaussian
When ρ = 0, this reduces to the independent case above.
- Two marginal Gaussian with a Gaussian copula
Starting from random variables with arbitrary marginals f1 and f2 (these are in this case two marginal normals) f1 ∼ 𝒩(μ1, σ1) & f1 ∼ 𝒩(μ2, σ2)
Mathematically this can be written as the two marginals
Y1 ∼ f1(θ1)
Y2 ∼ f2(θ2) To model the dependence we transform (Y1, Y2) into two uniforms through the probability integral transform. These are then back transformed into standard multivariate normal variables with correlation ρ this can be written in a single equation:
\[(z_1, z_2) = \begin{bmatrix} \Phi^{-1}\big(F_1(Y_1)\big) \\ \Phi^{-1}\big(F_2(Y_2)\big) \end{bmatrix} \sim \mathcal{N}\Bigg( \mathbf{0}, \begin{bmatrix} 1 & \rho \\ \rho & 1 \end{bmatrix} \Bigg), \quad F_i(\cdot) \text{ are the marginal CDFs}\]This chain of transformations shows how arbitrary marginal distributions can be linked through a Gaussian copula, with ρ controlling the dependence between the marginals.
Using LOO we show that the multivariate and copula Gaussian for all ρ are no different, but both being being better for all ρ != 0 than the two marginal Gaussians
Stan Implementations
We thus start with the stancode for the 3 models:
2 Marginal Gaussians:
data {
int<lower=1> N; // Number of observations
vector[N] y1; // Outcome variable 1
vector[N] y2; // Outcome variable 2
}
parameters {
vector[2] beta; // Coefficients for y2
vector<lower=0>[2] sigma; // Standard deviations for y1 and y2
}
model {
// Priors
beta ~ normal(0, 10);
sigma ~ normal(0, 5);
for(i in 1:N){
y1[i] ~ normal(beta[1], sigma[1]);
y2[i] ~ normal(beta[2], sigma[2]);
}
}
generated quantities{
vector[N] log_lik = rep_vector(0,N);
// Likelihood
for (n in 1:N) {
log_lik[n] += normal_lpdf(y1[n] | beta[1], sigma[1]);
log_lik[n] += normal_lpdf(y2[n] | beta[2], sigma[2]);
}
}
Multivariate Gaussian
data {
int<lower=1> N; // Number of observations
matrix[N, 2] y; // Observed outcomes [y1, y2]
}
parameters {
vector[2] beta; // Coefficients for y2
vector<lower=0>[2] sigma; // Standard deviations for y1 and y2
corr_matrix[2] Omega; // Correlation matrix
}
transformed parameters {
cov_matrix[2] Sigma; // Covariance matrix
Sigma = quad_form_diag(Omega, sigma);
}
model {
beta ~ normal(0, 10);
sigma ~ normal(0, 5);
Omega ~ lkj_corr(2); // LKJ prior on correlation matrix
// Likelihood
for (n in 1:N) {
y[n] ~ multi_normal(beta, Sigma);
}
}
generated quantities{
vector[N] log_lik;
// Likelihood
for (n in 1:N) {
log_lik[n] = multi_normal_lpdf(y[n] | beta, Sigma);
}
}
2 Marginal Gaussians and guassian copula:
functions {
real gauss_copula_cholesky_lpdf(matrix u, matrix L) {
array[rows(u)] row_vector[cols(u)] q;
for (n in 1:rows(u)) {
q[n] = inv_Phi(u[n]);
}
return multi_normal_cholesky_lpdf(q | rep_row_vector(0, cols(L)), L)
- std_normal_lpdf(to_vector(to_matrix(q)));
}
vector gauss_copula_cholesky_per_row(matrix u, matrix L) {
int N = rows(u);
int D = cols(u);
array[N] row_vector[D] q;
vector[N] loglik;
for (n in 1:N) {
q[n,] = inv_Phi(u[n,]);
loglik[n] = multi_normal_cholesky_lpdf(to_row_vector(q[n,]) |
rep_row_vector(0, D), L) - std_normal_lpdf(to_vector(to_matrix(q[n,])));
}
return loglik;
}
}
data {
int<lower=0> N;
matrix[N, 2] y;
}
parameters {
real<lower=0> sigma1;
real<lower=0> sigma2;
real<lower=0> mu1;
real<lower=0> mu2;
cholesky_factor_corr[2] rho_chol;
}
transformed parameters{
matrix[N, 2] u;
for (n in 1:N) {
u[n, 1] = normal_cdf(y[n, 1] | mu1,sigma1);
u[n, 2] = normal_cdf(y[n, 2] | mu2,sigma2);
}
}
model {
mu1 ~ normal(0, 10);
mu2 ~ normal(0, 10);
sigma1 ~ normal(0, 5);
sigma2 ~ normal(0, 5);
rho_chol ~ lkj_corr_cholesky(2);
y[, 1] ~ normal(mu1,sigma1);
y[, 2] ~ normal(mu2,sigma2);
u ~ gauss_copula_cholesky(rho_chol);
}
generated quantities {
real rho = multiply_lower_tri_self_transpose(rho_chol)[1, 2];
vector[N] log_lik; // per-trial log-likelihood
matrix[2,2] Sigma = multiply_lower_tri_self_transpose(rho_chol);
vector[N] ll_copula = gauss_copula_cholesky_per_row(u, rho_chol);
for (n in 1:N) {
// Marginal contributions
real ll_marg = normal_lpdf(y[n,1] | mu1, sigma1) + normal_lpdf(y[n,2] | mu2, sigma2);
// Total per-trial log-likelihood
log_lik[n] = ll_marg + ll_copula[n];
}
}
Fitting and Mutual Information
Now we simulate some multivariate data with correlation coefficient r, and then fit each of the stan models. Furthermore we can compare how well the stanmodels fit the data, but computing approximate leave one out crossvalidation (LOO-CV).
# Set up parallel plan
plan(multisession, workers = 10) # Adjust to your number of cores
# Correlations to simulate
r = seq(-0.6, 0.6, by = 0.05)
# Function to simulate and fit models for one correlation
simulate_and_fit <- function(cor) {
# Simulate data
n_out <- mnormt::rmnorm(n = 500, mean = c(1, 2),
varcov = cbind(c(1, cor), c(cor, 1)))
bdata <- data.frame(
y1 = n_out[,1],
y2 = n_out[,2]
)
# Marginal model
datastan_margin <- list(
y1 = bdata$y1,
y2 = bdata$y2,
N = nrow(bdata)
)
fit_margin <- marginal$sample(
data = datastan_margin,
refresh = 0,
adapt_delta = 0.99,
parallel_chains = 4
)
# Multivariate & copula models
datastan_multi <- list(
y = cbind(bdata$y1, bdata$y2),
N = nrow(bdata)
)
fit_multi <- multivariate$sample(
data = datastan_multi,
refresh = 0,
adapt_delta = 0.99,
parallel_chains = 4
)
fit_cor <- copula$sample(
data = datastan_multi,
refresh = 0,
adapt_delta = 0.99,
parallel_chains = 4
)
# Compute LOO comparison
loo_df <- data.frame(
loo::loo_compare(
list(
margin = fit_margin$loo(),
corpula = fit_cor$loo(),
multi = fit_multi$loo()
)
)
) %>% rownames_to_column()
loo_df$sim_correlation <- cor
return(list(loo_df,fit_margin,fit_cor,fit_multi))
}
# Run in parallel and combine results directly
loos <- future_map(r, simulate_and_fit, .progress = T)
Now we can plot the results of these model comparisons:
Plottiing the results of this
plot = map_dfr(loos,1) %>% filter(elpd_diff != 0) %>% mutate(models = rowname) %>%
ggplot()+
geom_pointrange(aes(x = sim_correlation, y = elpd_diff, ymin = elpd_diff-2*se_diff, ymax = elpd_diff+2*se_diff, col = models))+
theme_minimal()+
theme(legend.position = "top")+
theme(text = element_text(size = 24))+
scale_x_continuous(breaks = seq(-0.8,0.8,by = 0.1), labels = seq(-0.8,0.8,by = 0.1))
plot
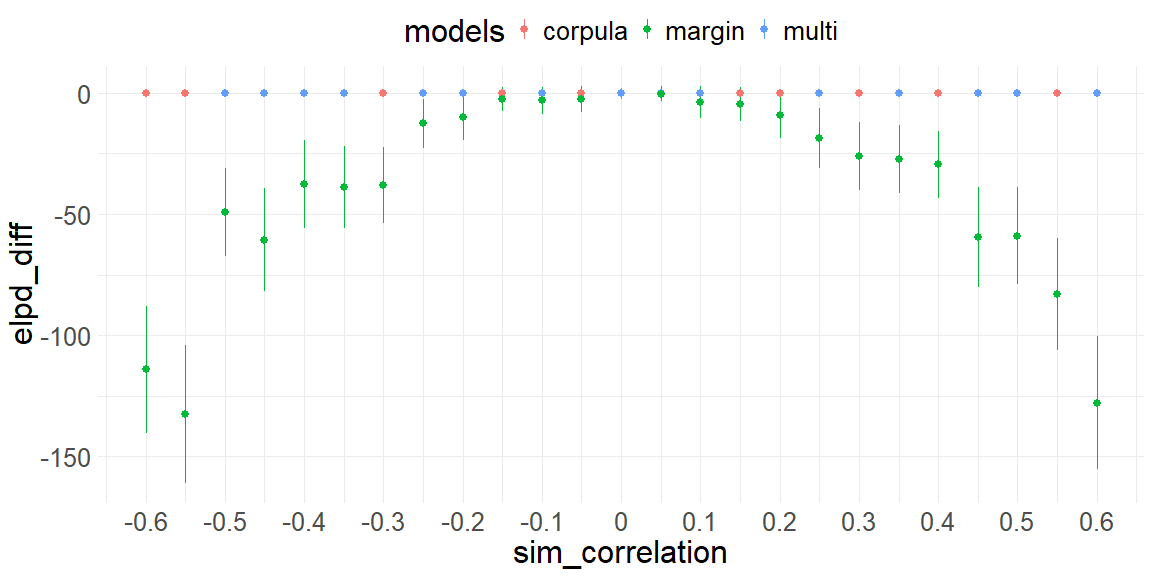
This figure shows the simulation results of 500 trials per simulation of correlation coefficients ((-0.6 ; 0.6 , by = 0.06) x-axis) of three models (colors) on their performance of LOO-CV. The plot shows how the marginal implementation explains the data worse in a quadratic way with respect to the simulated correlation coefficient. Furthermore, the copula and multivariate implementation share to explain the data the best (randomly alternating blue and red colors at elpd_diff = 0).
This shows that our implementation of the Gaussian copula is at least sensible because it behaves in exactly the same way as the multivariate normal. Further this simulation shows that the degree to which we explain the data worse with the marginal implementation is quadratic on the correlation coefficient. This is of course not a coincidence and is evident if we write the mutual information of our two random variables:
\[MI(x, y) \leq \frac{1}{2} \log \left( \frac{1}{1 - r^2} \right)\]this quantity is measured in bits. The interesting part is that it depends on the square of the correlation coefficient. Which aligns with the plot above, when plotted ontop with a scaling factor of the sample size we see:
plot+geom_line(data = data.frame(),aes(x = r, y = (-1/2*log(1/(1-r^2)) * 500)))
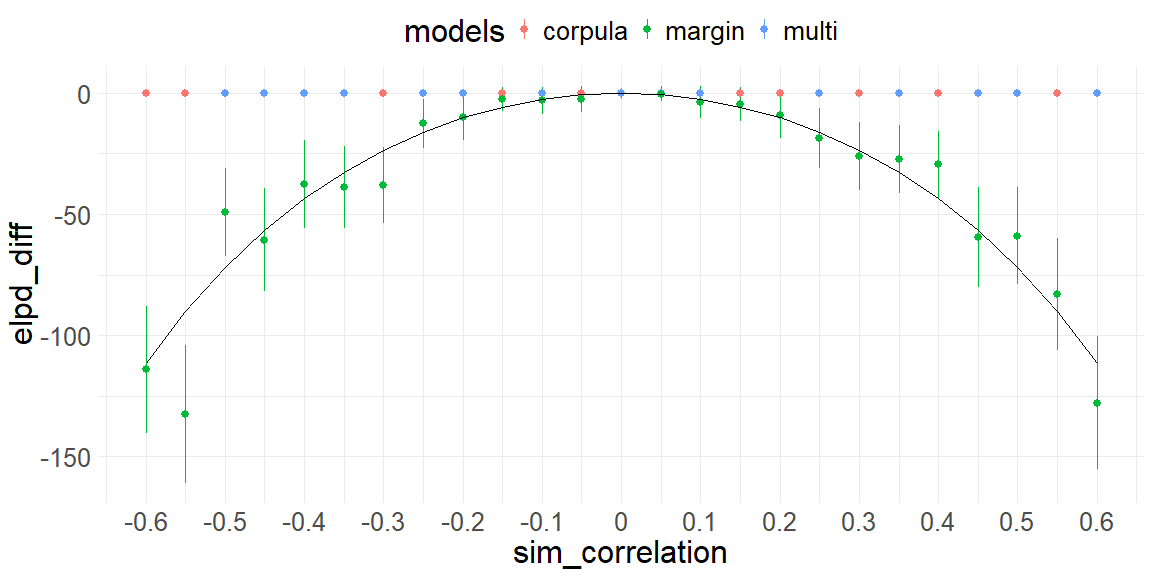
In short, we can implement and showcase that the copula implementation works when comparing guassian distributions and correlation coefficients and guassian copulas and that the LOO-CV captures the underlying relationship. Next, the strength of the copula implementations will be showcased, that is when the two marginals aren’t Gaussian.
